PgxSAVy About
PgxSAVy is a tool for quality control and annotation of variant peptides after FDR in proteogenomics.
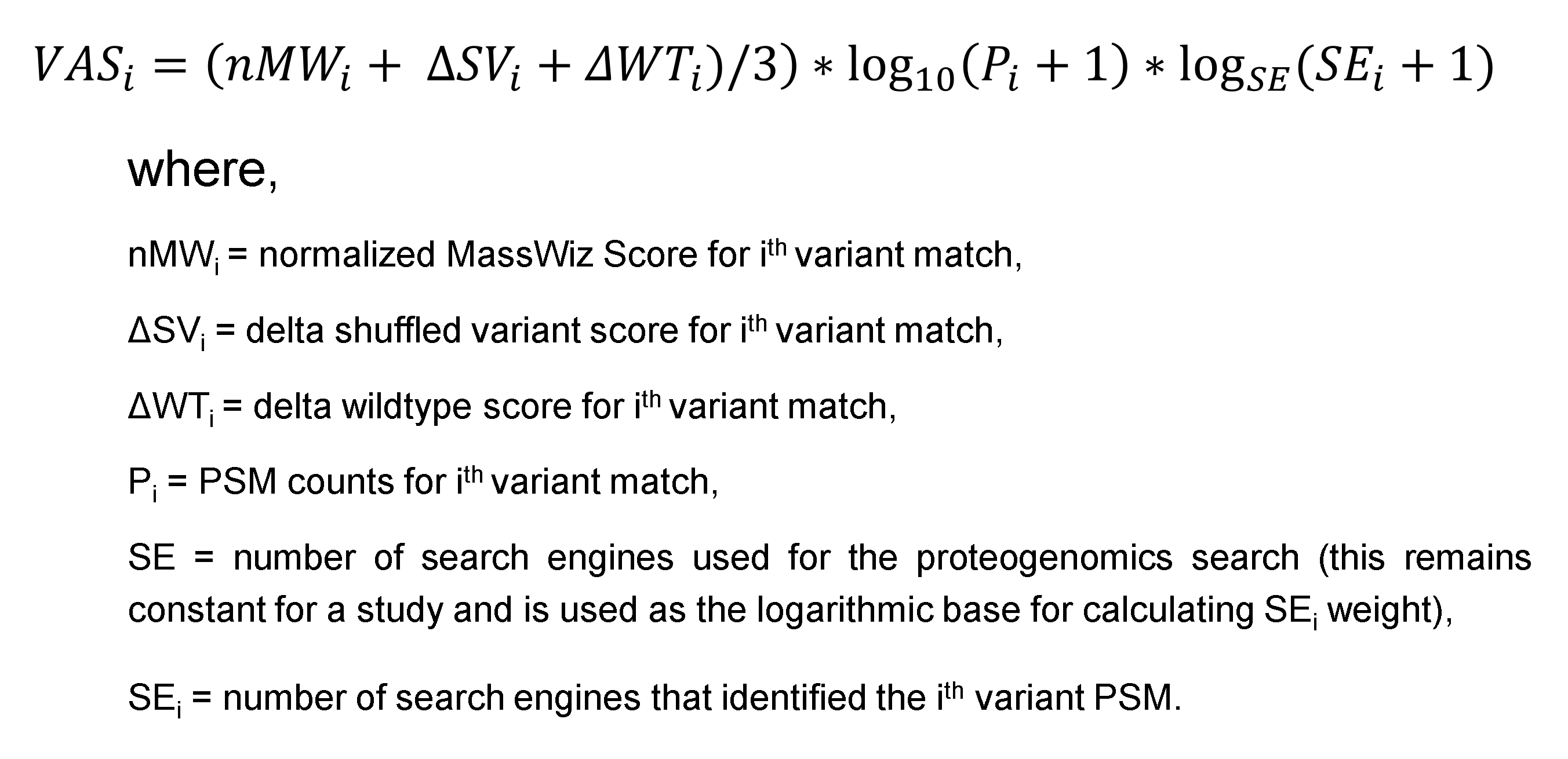
The PgxSAVy tool is broadly divided into:
- Data layer: This represents the data files supported for various algorithms. This layer reads the file formats and converts them to easily handled text formats (TSV/CSV) for faster, easier access and quick filtering for results parsing later. MS/MS data is required in 'MGF' format for spectra level information.
- Input Layer: Input layer has file parsers to convert from the native result formats to tabular formats. The file reading function then reads the spectrum, peptide, variant, modifications and protein details. Based on the spectrum title, it extracts information from MS/MS data.
- Rescoring Layer: Based on the VAS scoring method, for each PSM, a normalized MW score is calculated for variant peptide, corresponding wildtype peptide and shuffled variant decoy peptides. These calculated scores are saved in a two-dimensional array and zscore and p-value is estimated. For estimation of zscore, negative scoring PSMs are used to create a null hypothesis for normal distribution. If PSM fails to follow the normal distribution, it belongs to a uniform distribution which reflects the confident PSMs.
- Annotation Layer: The p-value filters (≤ x, where x is desired cut-off) can be directly applied to classify variant peptides as confident, semi-confident and doubtful. Further, isobaric and disease annotation is done over the identified variants.
The following flowchart shows the broad details of the PgxSAVy tool functionality and data flow during rescoring and annotation.
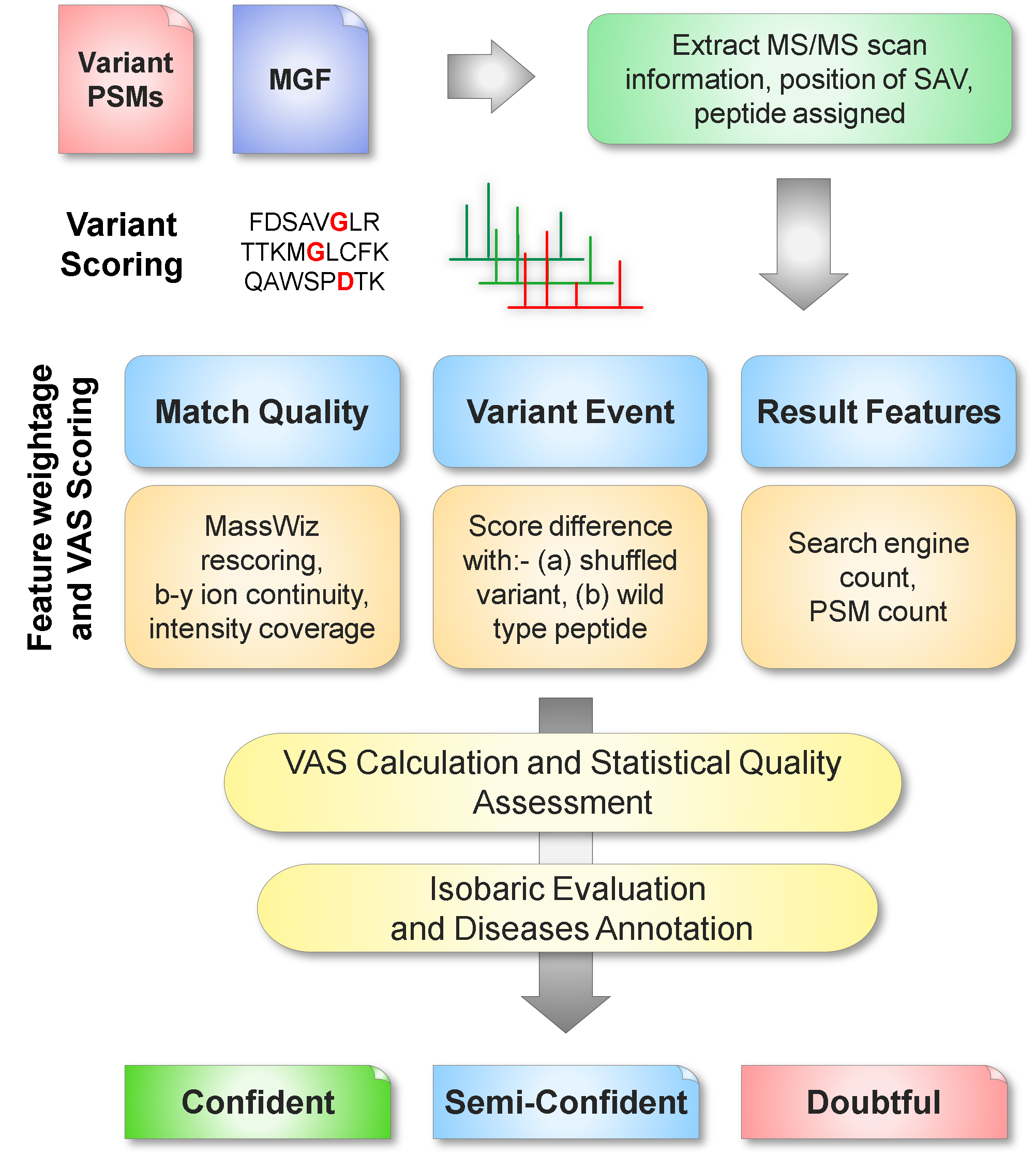
Rescoring formulae/methods in PgxSAVy
The PgxSAVy tool supports the following formulations on rescoring.
VAS is defined as --